The reviewer has made very common but tragic mistakes. There are many papers showing extreme problems with all the methods the reviewer likes. May need to go to a journal that has qualified statistical reviewers. Start with papers by Margaret Pepe. Bad power from studying changes in c-index can be easily simulated; can’t put my finger on a paper. But if you keep trying that journal you might mention that the original developer of the c-index backs you up.
Thank you very much for your reply. I will definitely start with the Pepe ones. I wish all journals had qualified statistical reviewers…
Do you perhaps have a good reference to cite, where is stated that p-values for differences in c-statistics and p-values for NRI and IDI are problematic? Would really much help me.
I am currently trying to familiarise myself with the literature on this, but as I only have a layman’s background in statistics, this is of course very challenging.
I’m sure someone can find such a reference for us. Think about it this way: Differences in c equate to differences in Wilcoxon statistics. If doing Wilcoxon tests to compare A with B and B with C no one computes the difference in two Wilcoxon stats to compare A with C. That would detect weird alternatives and have poor power.
Ugh… I feel for you Julia.
@f2harrell giving permission to name drop him is great!
Margaret Pepe’s papers about NRI:
-
Pepe MS, Fan J, Feng Z, et al. The Net Reclassification Index (NRI): A Misleading Measure of Prediction Improvement Even with Independent Test Data Sets. Stat Biosci 2015; 7: 282–295.
-
Pepe MS. Problems with risk reclassification methods for evaluating prediction models. Am J Epidemiol 2011; 173: 1327–1335.
I think Jorgen Gerds wrote about the IDI and issues when there was miscalibration of models.
Julia, I defer to the world class experts here but there are clinical considerations which I have learned over the past 4 decades of studying the clinical behavior of relational time series matricies of biomarkers, matrix patterns and derivatives. These affect the validity of the statistics.
It is first important to consider:
- What type of biomarker(s) you are investigating?
Biomarkers may be derived directly from the condition under test (hcg) or indirectly from a complication (creatinine), a general mortality signal (lactate) or a physiologic response to the condition (WBC)?
- What type of condition you are investigating?
A well defined disease or a broadly defined synthetic syndrome comprised of many diseases?
-
What is the volitility of the biomarker. (what is the derivative range of the biomarker with and without the condition under test )?
-
What factors affect the biomarker volitility and is this volitility a function of the condition?
-
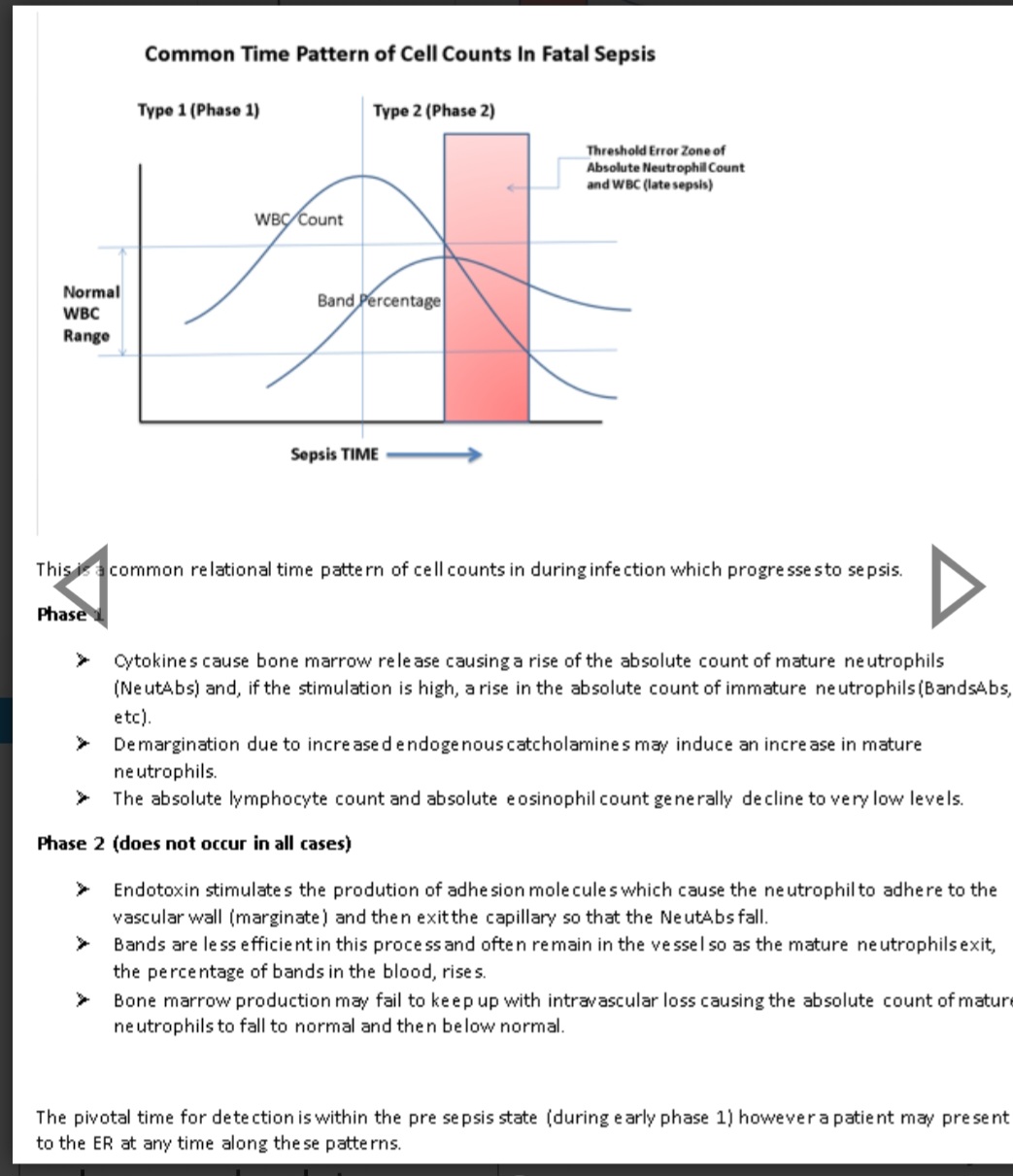
Is the biomarker depletable or biphasic. (WBC is a depletable biomarker often high early and low late)?
6 Is the biomarker a late marker such that it is generally normal early in the disease (eg bands, lactate)?
-
Does the biomarker have a high normal phenotypic range. (eg platelets)?
IMHO, these are pivotal considerations and it is intuitive that these account for much of the failure of AUROC as a stand alone biomarker test.
In an example, bands (a type of immature neutrophil), were abandoned in many hospitals because biomarker studies demonstrated that the absolute neutrophils had a better AUC. However both are “response biomarkers”, moderately volatile biomarkers, and bands are late markers whereas absolute neutrophils are both early markers and depletable.
Had the authors of these studies engaged in due considerations of the above relational times series matrix factors they would not have determined that absolute neutrophils were superior to (and could replace) bands.
The loss of bands as a biomarker in many hospital systems remains and has echoed in the halls of the ICU as a function of very late detection of life-threatening infection for decades.