Hi all, I asked this on Cross-Validated (https://stats.stackexchange.com/questions/362482/equation-for-standard-error-of-linear-predictor-and-95-prediction-intervals-fro) but I figured this would be an even better place.
I would like to present 95% prediction intervals in an online risk calculator.
-
After fitting a logistic model with
lrm(which includes some restricted cubic splines), I export the equation usinglatex()and program the model as a risk calculator. -
The trouble is that I want to also find the equations for the upper and lower limits of the 95% prediction intervals around the predicted probabilities to help users see the uncertainty in the predictions.
-
I know how to produce prediction intervals using the
predict(model,data,type="lp",se.fit)method, but it is not the actual values I am interested in. Rather I need to get the equation.
The scenario is a common one in risk modelling but risk calculators very rarely show upper and lower prediction intervals.
How to calculate standard errors of the linear predictor?
From Sofroniou N, Hutcheson GD. Confidence intervals for the predictions of logistic regression in the presence and absence of a variance-covariance matrix. Understanding Statistics: Statistical Issues in Psychology, Education, and the Social Sciences. 2002 Feb 2;1(1):3-18.
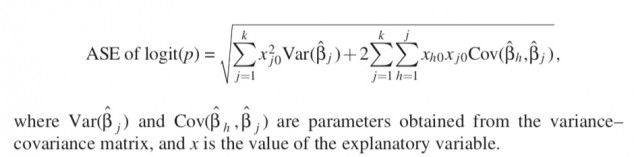
Not sure how I can create a single equation in the form logit§ = B1X1+1.96ASE_of_B1 + B2X2+1.96ASE_of_B2… that I can then work with in excel or javascript etc to give people upper and lower 95% prediction limits.
Initially I thought I could just do something like:
#the main lrm object
fit<-lrm(y~rcs(x1,3)+x2+x3,data=data,X=T,Y=T)
#just creating some lrm objects to update later for user with latex()
fit.ucl<-fit
fit.lcl<-fit
#update the coefficients in the lrm objects to represent the upper and
#lower bounds of the confidence intervals
fit.ucl$coefficients<-(fit$coefficients + 1.96*(sqrt(diag(vcov(fit)))))
fit.lcl$coefficients<-(fit$coefficients - 1.96*(sqrt(diag(vcov(fit)))))
#output the equations for each in latex
latex(fit)
latex(fit.ucl)
latex(fit.lcl)
but this is using only the diagonal of the vcov matrix and doesn’t take into account the off-diagonals (the covariances) as the above equation requires.
My model also includes restricted cubic spline-transformed variables, the interpretation of which is greatly facilitated by latex(model) or Function(model). I could write an equation to compute SE of the LP in real time using the full varcov matrix if I could easily convert the b coefficients on the splinevar, splinevar` notation into b* splinevar + b* pmax(splinevar-knot1,0)^3 - b* pmax(splinevar-knot2,0)^3 + b* pmax(splinevar-knot3,0)^3.
Thanks for considering,
Pavel
