Hi all.
I’m running a survival model (a cox regression) - on a large dataset of ~29,000 patients who have had a diagnosis, and we are monitoring their time to survival from this diagnosis.
This is real data, and unpublished, so I’m being slightly vague. There is a biomarker, B, that we have measured (often repeatedly) prior to the diagnosis, D, and therefore there are multiple potential approaches to defining this biomarker:
Most recent test prior to diagnosis, D (minimise B-D)
Maximal test result
Minimal test result
Mean test result
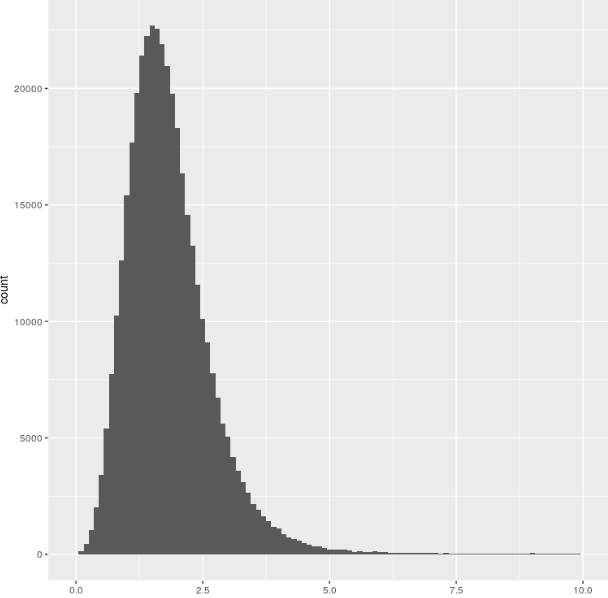
Here’s the distibution of B (all recorded tests - 350,000!)
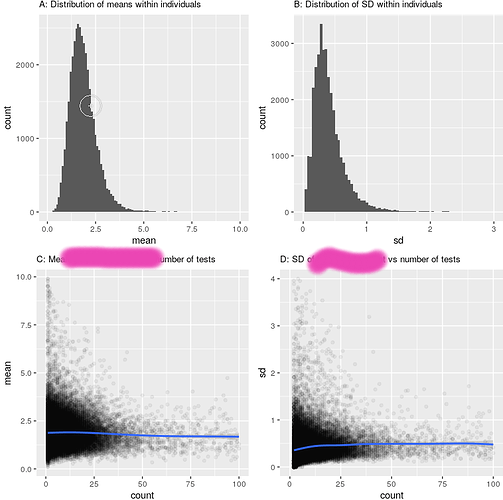
So, fairly normally distributed, and there isn’t a strong relationship to testing strategy:
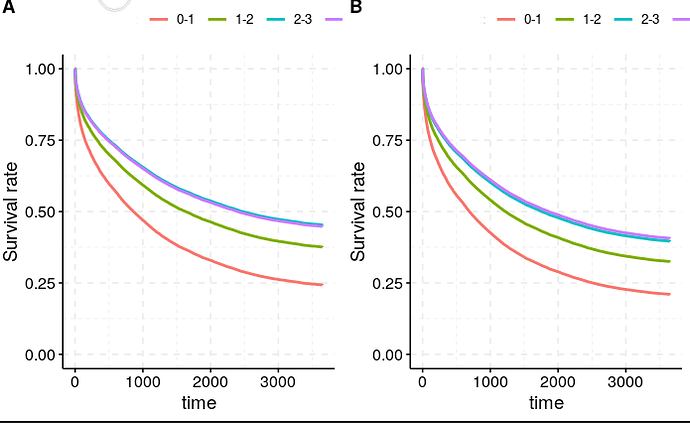
The Cox regression result is pretty clear: there is a strong relationship between a low level of this biomarker and survival (HR 0.89, p<10^-30). This is born out by both testing strategies (A : minimum appoach, B: maximum approach - adjusted for relevant covariates using ggadjust - the unadjusted look similar but don’t work with cowplot/gridarrange!)
Now the relationship seems pretty robust to how we measure this Biomarker, and which strategy we take (left is minimum, right is maximum approach), despite the actual figures being quite different (mean B across the population 1.53 for the ‘minimum’ approach, 2.53 for the maximum approach).
To plot the curves above, i split the biomarker into 4 groups : <1, 1-2, 2-3, >3 (see the distribution above). This was simply to plot nearly. Unsuprisingly, changing the definition changed the composition of the groups (there were many more people who had a minimum B of less than 1, than those who had a maximum B of less than 1). However, the median survival did not budge
Look at the Kaplan-Meir fits here:
Maximum ever B:
Call: survfit(formula = Surv(censor_time, censor) ~ categorised_B,
data = df)
1 observation deleted due to missingness
n events median 0.95LCL 0.95UCL
categorised_B=0-1 1021 821 613 497 737
categorised_B=1-2 11112 7663 1182 1134 1237
categorised_B=2-3 11095 6547 1903 1806 2024
categorised_B=>3 5425 3202 1835 1714 1997
Minimum B:
> Call: survfit(formula = Surv(censor_time, censor) ~ categorised_B,
> data = df)
>
> 1 observation deleted due to missingness
> n events median 0.95LCL 0.95UCL
> categorised_B=0-1 6937 5278 720 681 766
> categorised_B=1-2 15869 9874 1661 1587 1724
> categorised_B=2-3 4896 2568 3010 2733 3321
> categorised_B=>3 951 513 2794 2371 3334
I don’t get this. How come the median survivals are so similar - these are actually remarkably different groups? Am i overthinking this?
I would have thought (given that having a low B is bad), having a Max B less than 1 would be much worse than having a min B less than 1, as there is simply fluctuation around the mean.
Any thoughts (or explanations that this is not really interesting) useful - I am a clinician not a statistician!
Gus