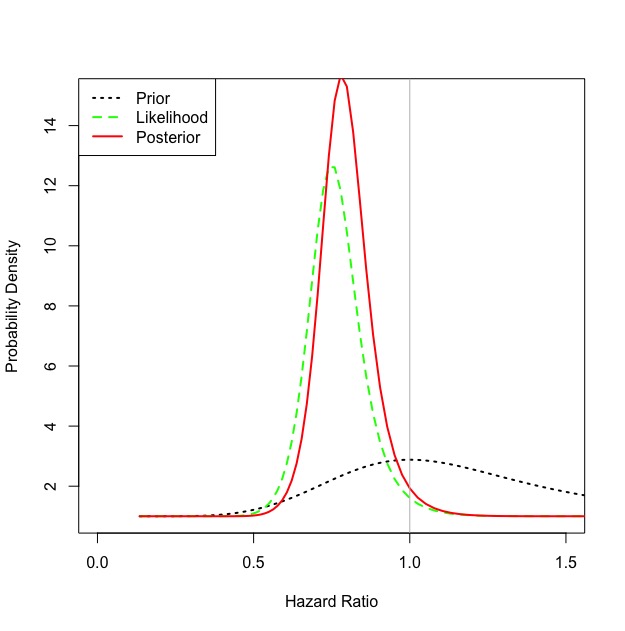
I estimated the Bayesian posterior using a skeptical prior (mean no effect, standard deviation extending 5% past MCID) and methods proposed in this paper. As you suggested the probability of HR<1 was 0.95, with a HR of 0.78 95% Certainty Interval 0.59-1.04.
I’d argue this is strong evidence for adopting the capillary refill time as the target end-point, instead of the more invasive and less direct measure of tissue hypoperfusion given by lactate.
Limitation: This method assumes normal distributions of the parameters.
R Code (Please point out if there are any errors so I can use again)
*Update: Code has been refined with help from @BenYAndrew
#Calculating MCID#
#Here I am using the estimated reductions from the power calculation to get an OR for the MCID (may need to be converted to RR instead of OR)#
n < - 420 #Sample size
a <- 0.3 * n #Intervention and Outcome
b <- 0.45 * n #Control and Outcome
c <- n - a #Intervention No Outcome
d <- n - b #Control No Outcome
MCID <- ((a+0.5) * (d+0.5))/((b+0.5) * (c+0.5))
#Hazard Ratio
HR <- 0.75
UCI <- 1.02
#Calculate Prior
#Skeptical prior mean estimate
#Calculating skeptical prior SD estimate for 5% probability of exceeding projected estimate
z <- qnorm(0.05)
prior.theta <- log(1)
prior.sd <- (log(MCID)-prior.theta)/z
#Enthusiastic Prior
prior.theta <- log(MCID)
prior.sd <- (log(1.05)-log(MCID))/qnorm(0.975)
#Calculate Likelihood
L.theta <- log(HR)
L.sd <- (log(UCI)-log(HR))/qnorm(0.975)
#Calculate Posterior
post.theta <- ((prior.theta/prior.sd^2)+(L.theta/L.sd^2))/((1/prior.sd^2)+(1/L.sd^2))
post.sd <- sqrt(1/((1/prior.sd^2)+(1/L.sd^2)))
#Calculate posterior median effect and 95% certainty interval
cbind(exp(qnorm(0.025, post.theta,post.sd, lower.tail = T)), exp(qnorm(0.5, post.theta,post.sd)), exp(qnorm(0.975, post.theta,post.sd)))
#Calculate probability benefit (HR < 1.0)
pnorm(log(1), post.theta,post.sd, lower.tail=T)
# Plot the prior density
mu.plot <- seq(-2,2,by=0.025)
prior <- dnorm(mu.plot, prior.theta, prior.sd)
likelihood <- dnorm(mu.plot, L.theta, L.sd)
posterior <- dnorm(mu.plot, post.theta, post.sd)
plot(exp(mu.plot), exp(prior),
type=“l”, col=“black”, lty=3,
xlim=c(0,1.5),
ylim=c(1,15),
lwd=2,
xlab=“Hazard Ratio”,
ylab=“Probability Density”)
#likelihood
lines(exp(mu.plot), exp(likelihood), col=“green”,lwd=2,lty = 2)
#posterior
lines(exp(mu.plot), exp(posterior), col=“red”,lwd=2)
abline(v=1, col = “gray”)
legend(“topleft”,
col=c(“black”,“green”,“red”),
lty=c(3,2,1),
lwd=2, #line width = 2
legend=c(“Prior”, “Likelihood”, “Posterior”))