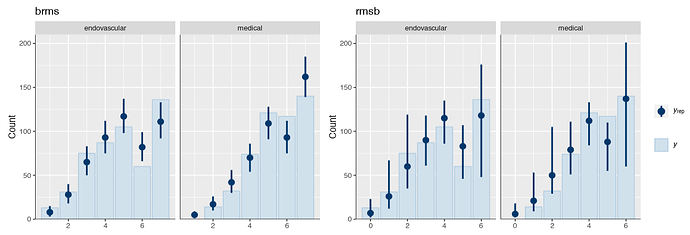
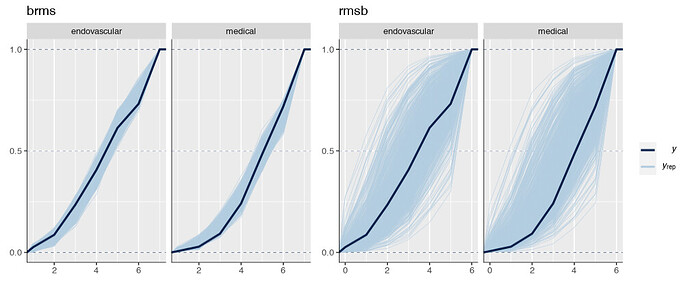
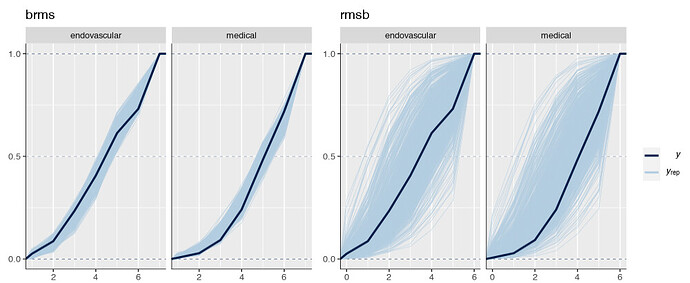
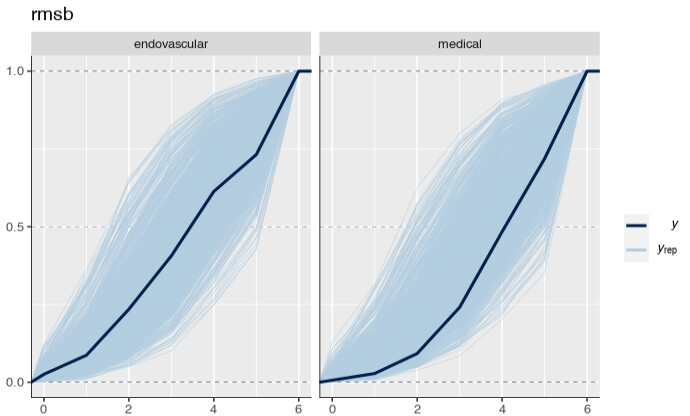
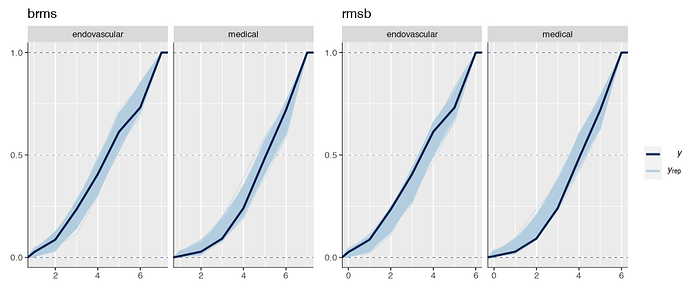
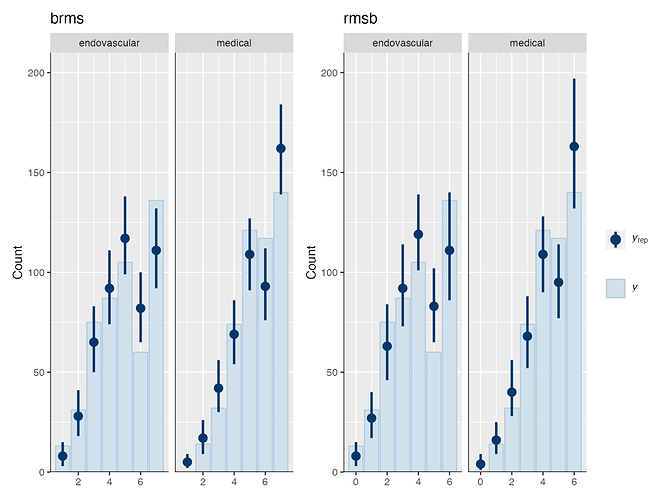
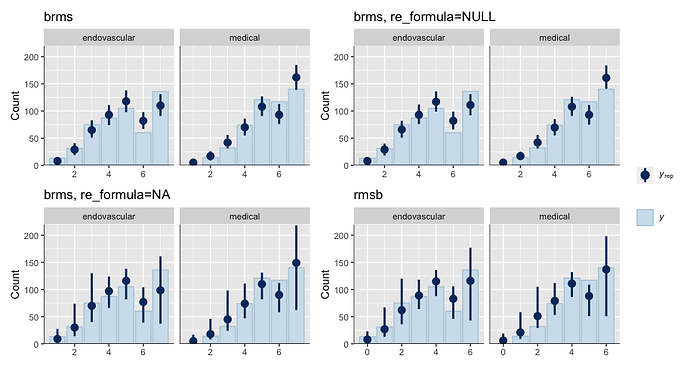
Is it possible that the pp_check implementation for the rmsb model is treating the random effects differently than the brms pp_check implementation when generating model predictions? For example, the code below reproduces your p1 and p2 plus two additional plots: p1a sets re_formula=NULL, which includes uncertainty due to random effect group levels, while p1b sets re_formula=NA which excludes uncertainty due to random effect levels. The re_formula argument is passed on to posterior_predict, and re_formula=NULL is the default. The plots are below, followed by the code.
In the top row, the plot with an explicit re_formula=NULL is the same as the original brms plot, which is expected, given that the posterior_predict default is re_formula=NULL. In the bottom row, the plot of the brms model with re_formula=NA is similar to the plot generated from the rmsb model (though maybe this is just coincidental).
I’m actually confused by this. I expected the plot with re_formula=NULL to have wider uncertainty bands than the plot with re_formula=NA, because the former includes uncertainty due to varying levels of the random intercept while the latter does not. In any case, I thought I’d post this in case it has something to do with the differences you’re seeing between the two posterior checks.
p1 = pp_check(PO_brms, group = "x", type = "bars_grouped", ndraws = 1000) +
labs(title = "brms") + coord_cartesian(ylim = c(0, 220))
p1a = pp_check(PO_brms, group = "x", type = "bars_grouped", ndraws = 1000,
re_formula=NULL) +
labs(title = "brms, re_formula=NULL") + coord_cartesian(ylim = c(0, 220))
p1b = pp_check(PO_brms, group = "x", type = "bars_grouped", ndraws = 1000,
re_formula=NA) +
labs(title = "brms, re_formula=NA") + coord_cartesian(ylim = c(0, 220))
p2 = pp_check(PO_rmsb, group = "x", "bars_grouped", ndraws = 1000) +
labs(title = "rmsb") + coord_cartesian(ylim = c(0, 220))
p1 + p1a + p1b + p2 + patchwork::plot_layout(guides = 'collect')