There was a community challenge for predicting response to anti-PD1 response prediction a few years ago. The results of the challenge was later published.
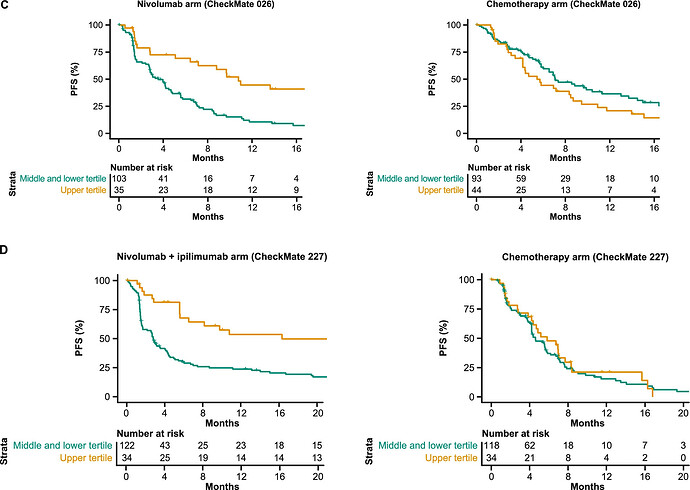
Not all patients with NSCLC achieve a response with ICIs. Consequently, there is a strong need for predictive biomarkers of outcomes with ICIs [9]. Studies reporting associations with ICI response in NSCLC have been limited by small sample sizes from single ICI treatment arms [17, 19, 20]. This Challenge addressed these shortcomings by using two large and well-characterized phase III RCTs and by comparing predicted responses between ICI- and chemotherapy-treated arms, thereby distinguishing treatment response prediction from prognostic effects.
The thing to be noted here is that they evaluated response prediction models from public datasets by applying them separately to each treatment arm of 2 clinical trials! Theoretically this seems very problematic to me because we know that treatment arms might not be identical at all due to complex treatment effects, drop-out patterns, side effects and so forth. The fact that the operating characteristics look different per arm is a very crude form of validation. What are all the ways that treatment arms might differ (for reasons only indirectly related to treatment assignment)? What are the known violations one might expect in trials like Checkmate? Do people in the field understand this?
Am I being too harsh in thinking this paper doesn’t seem to understand the fundamental problems of evaluating predictive biomarkers?