Piggybacking on this question.
I am interested in visually inspecting whether a variable is linearly associated with survival in a time to event model. Based on my understanding, fitting a naïve model without any transformation of a continuous variable, the hazard ratio will be an estimate of the effect on the hazard when increasing the variable by one unit. If the variable is log2-transformed, the hazard ratio will be an estimate of the effect when doubling the variable.
Both these analyses should assume (1) proportional hazards over time as well as (2) that the hazard rate is proportional over varying levels of the variable. It is the second point I’m interested in here, i.e. whether I can assume that the effect (on the hazard rate) of “increasing” my (untransformed) variable of interest from 10 to 20 is similar to increasing it from 20 to 30. For a log2-transformed variable, if I understood correctly, one would be interested in whether an increase from 10 to 20 would be similar to increasing it from 20 to 40.
I am trying to use real data to explore functional forms, but struggle to find out whether I got the code right (see below) to plot the hazard rate vs a continuous variable (univariable model).
# Histogram of the marker’s distribution
p1 <- df %>%
ggplot(aes(x = marker))+
geom_histogram(bins = 50)+
theme_dca()+
labs(title = "Histogram of marker values, N=270")
# Untransformed
fit <- cph(Surv(time, endpoint) ~ marker, df)
p2 <- ggplot(Predict(fit, marker = seq(min(df$marker), max(df$marker), length = 200))) +
#scale_x_continuous(limits = c(0, 300))+
labs(title = "Untransformed")
# Log2-transformed
fit <- cph(Surv(time, endpoint) ~ log2(marker), df)
p3 <- ggplot(Predict(fit, marker = seq(min(df$marker), max(df$marker), length = 200))) +
scale_x_continuous(limits = c(0, 300))+
labs(title = "Log2")
fit <- cph(Surv(time, endpoint) ~ rcs(marker,4 ), df)
p4 <- ggplot(Predict(fit, marker = seq(min(df$marker), max(df$marker), length = 200))) +
scale_x_continuous(limits = c(0, 300))+
labs(title = "RCS 4 knots")
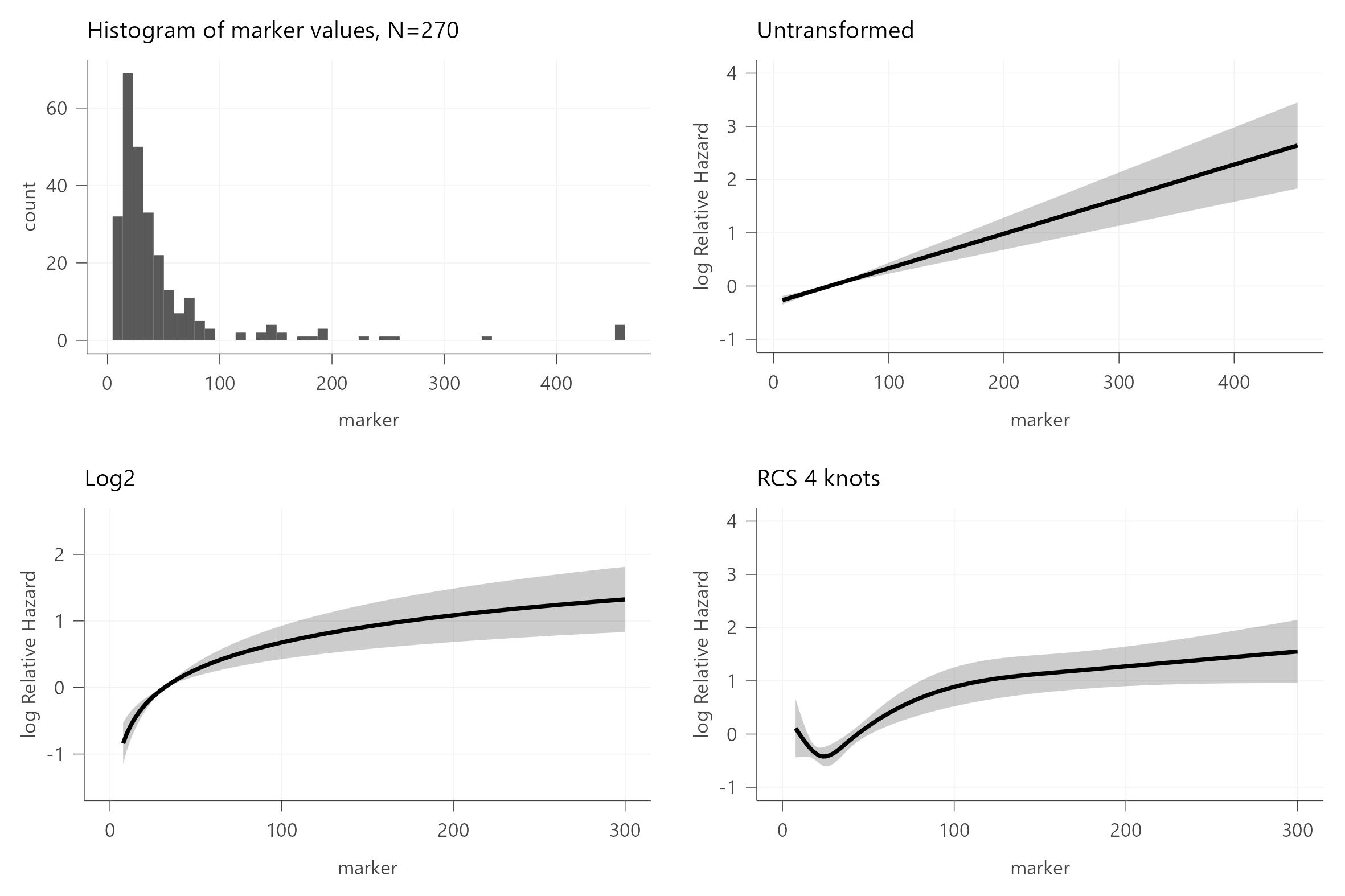
I understand from Harrell’s post above that fitting flexible functions like rcs() is useful – but (given that my code above is correct) I am not entirely sure how to move forward. I think both the log2 and rcs-transformed data indicate a departure from linearity, particularly at low concentrations. (Since this is a circulating analyte this is hardly surprising.) But does this mean that the hazard ratio from my log2 model cannot be directly interpreted? The RCS model also suggests departure from linearity, but as far as I’ve understood this transformation is flexible enough to handle non-linear relationships – so what would be the natural next step in my case? And I suppose the interpretation would be conditional on (omitted) covariates that may confound or interact with the variable?
