If have optimised the computational time by an order of magnitude by parallelising. It now takes roughly 10 minutes. For anyone interested:
library(foreach)
library(doParallel)
library(doRNG)
outer_B ← 2000
inner_B ← 10
my.cluster ← parallel::makeForkCluster(7)
doParallel::registerDoParallel(cl = my.cluster)
doRNG::registerDoRNG(seed = 1) # each fork of each iteration is independent and reproducible
val_metrics_1 ← foreach(i = 1:outer_B) %dopar {
data1 ← as.data.frame(
impute.transcan(
imputation,
data = data,
imputation = sample(imputation$n.impute, 1),
list.out = TRUE,
pr = FALSE,
check = FALSE
))
mod1 ← lrm(
formula = outcome ~ covariates,
data = sample(data1, nrow(data1), replace = TRUE), x = TRUE, y = TRUE)
validate(mod1, kint = 1, B = inner_B)}
parallel::stopCluster(cl = my.cluster)
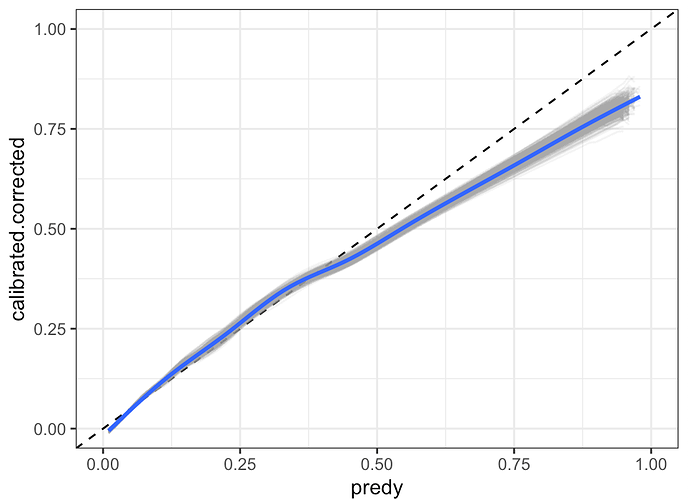
I also have a parallelised solution to extract confidence intervals around an optimism-corrected calibration curve that includes the variability due to both imputation and bootstrap resampling.