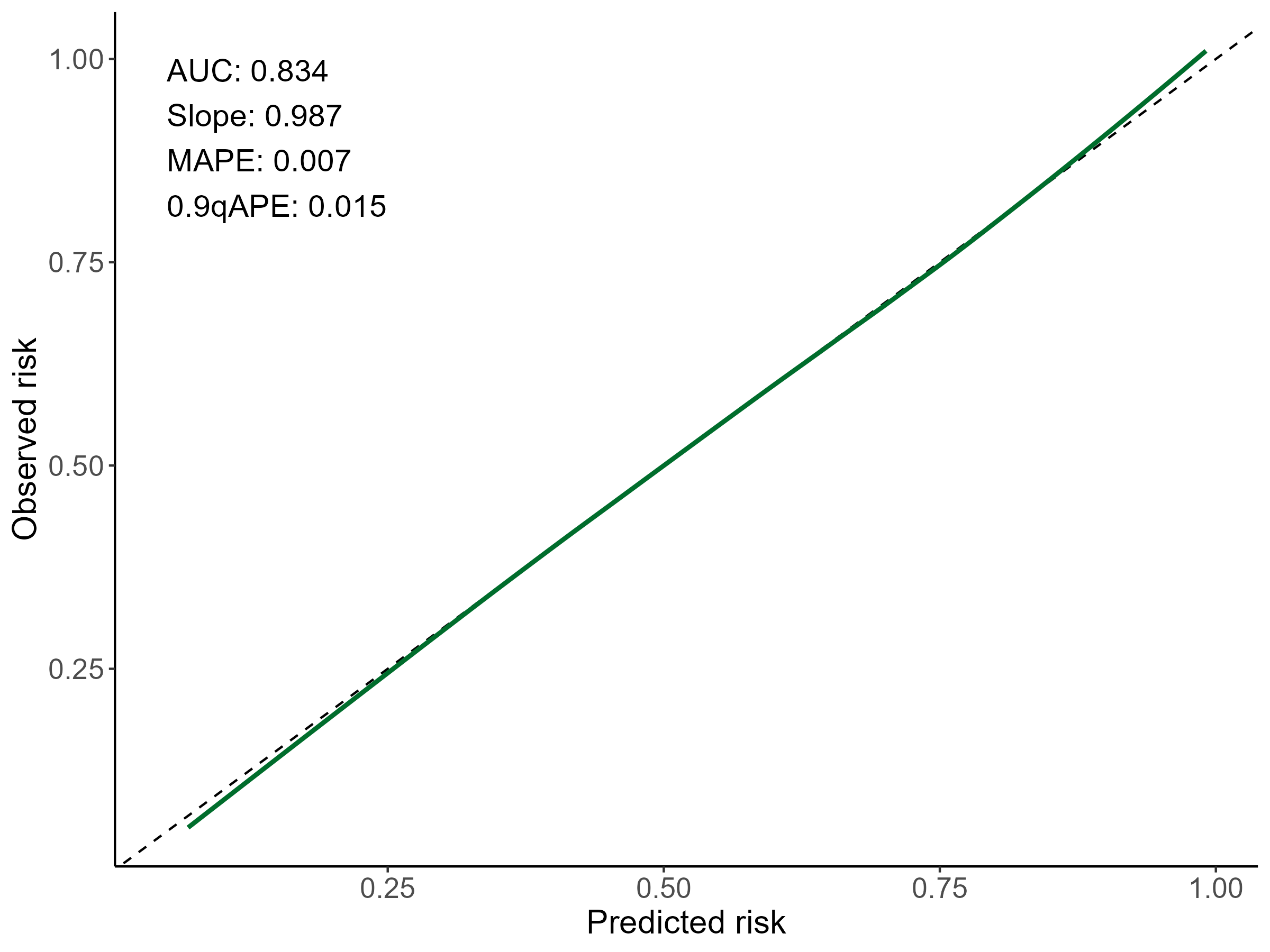
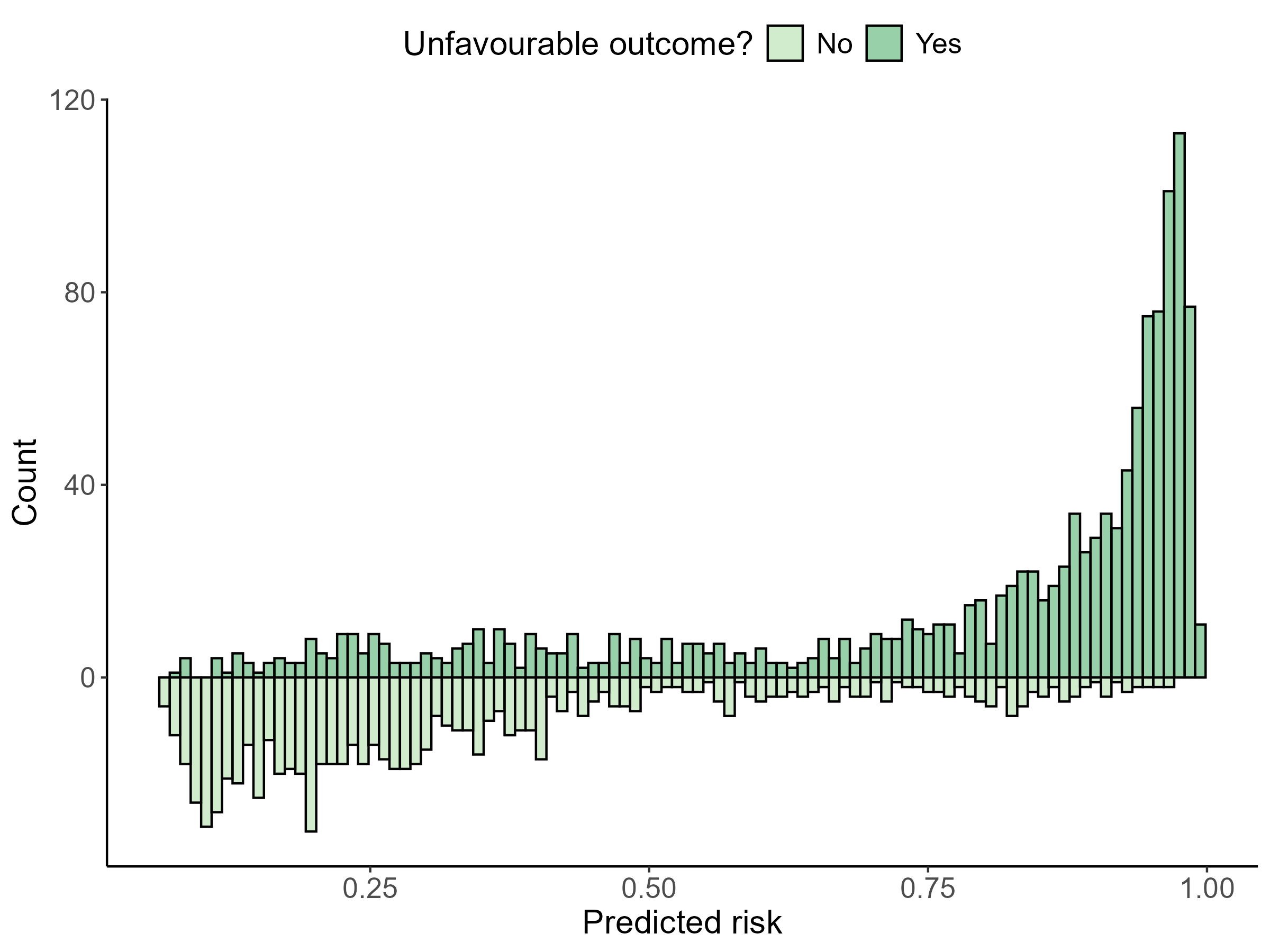
Thank you very much, Dr. @FurlanLeo and Dr. @f2harrell.
I have moved the post here, and if anything I do is inappropriate, please let me know.
First, I want to confirm whether my understanding in the aforementioned post is totally correct, except for the part about the Test sample.
Next, I want to briefly describe my understanding of the Efron-Gong Optimism Bootstrap. (I have the rms book, but for me, I still find many contents difficult to understand, so I would like to get your confirmation to ensure I am not doing anything wrong)
Assuming that there is a model y ∼ k * x, where x is a predictor and k is the coefficient of x, and y is the outcome.
- Calculate the performance measures (e.g., R2) in the original data, which corresponds to the performance measures in the
Original Samplecolumn inrms.validate. - Calculate the performance measures in a bootstrap sample, and there is a new coefficient
k1(model:y ∼ k1 * x) in this bootstrap sample. - Use the coefficient
k1(model:y ∼ k1 * x) to get new performance measures in the original data. - Calculate the optimism by (performance measures in step 2) - (performance measures in step 3).
- Repeat steps 2 to 4 for
ntimes to getnperformance measures in both bootstrap samples and original samples. - Calculate the average value of the performance measures in step 5, which correspond to the performance measures in the
Training Sample(average in step 2) andTest Sample(average in step 3) columns respectively inrms.validate. - Calculate the average value of the optimisms in step 6, which corresponds to the performance measures in the “Optimism” column in
rms.validate.
ie. Training Sample - Test Sample = Optimism
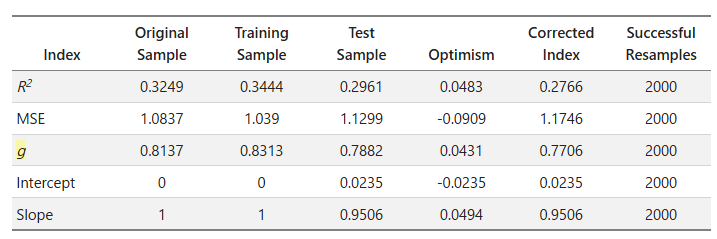
Thank you for your patience, and I really appreciate it if anyone could point out any misunderstandings I have. It would be very helpful for me.