Hi Prof Harrell,
Thank you for your great R package.
I have some problems using the rms package.
- “datadist” in subgroup analysis;
When performing a subgroup analysis, is it advisable to create a new “datadist” object?
For example, when performing a subgroup analysis only in male, should I do create a new “datadist” object specific to that subgroup?
ddist ← datadist(dt)
options(datadist = ‘ddist’)
cph(Surv(py, d) ~ rcs(AGE, 4),data = dt)ddist.m ← datadist(dt[sex = m])
options(datadist = ‘ddist.m’)
cph(Surv(py, d) ~ rcs(AGE, 4),data = dt[sex = m])
-
Use of “rcs”;
Should “rcs” be used for all continuous variables? If not, how can I determine when to use “rcs”?
For example, by observing plots of rcs or changes in AIC or likelihood ratios? -
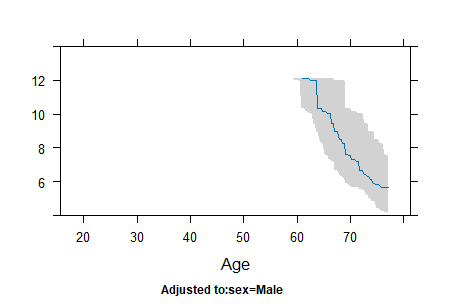
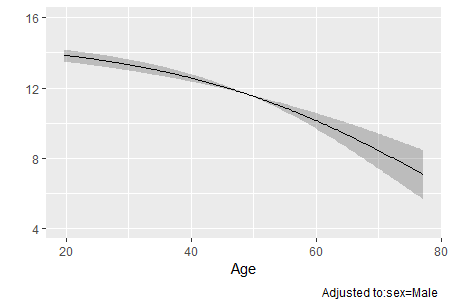
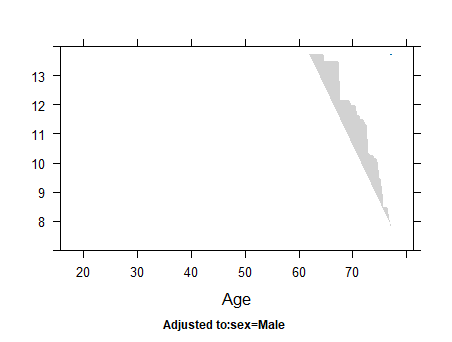
Horizontal axis in plots of rcs;
In my data, the blood glucose range is from 0 to 100 mg/dL, but there are values above 100 mg/dL on the x-axis. I’m not very familiar with statistics; is the x-axis in the rcs plot a simulated value? It doesn’t seem to correspond directly to the actual values.On my rcs plot, there is a steep increase in the hazard ratio for values above 100 mg/dL . Therefore, I suspect that the risk values for this part are also speculative. Am I understanding this correctly?
I apologize for taking up your time with these simple questions; I am still learning.
Thank you.
li