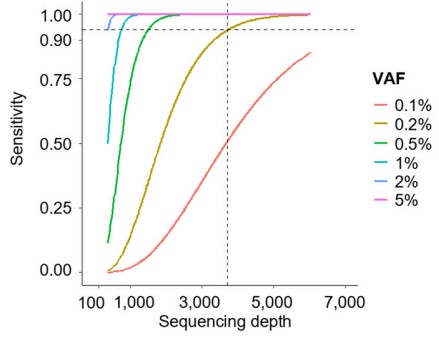
Figure Legend: Assay sensitivity for the detection of CH-related variants at different variant allele fractions (VAF). Dashed lines show the median sequencing depth of our study (3,712x; vertical line), and the sensitivity to detect CH variants at the minimum VAF threshold (0.2%) used to define CH (horizontal lines)
Hi,
I would like to perform a sensitivity analysis similar to the one shown in the attached image, which illustrates the sensitivity of detecting variants at different variant allele frequencies (VAF) as a function of sequencing depth.
Could you advise on the type of input data required for this analysis, as well as the corresponding R code needed to perform it?