Thanks @scboone for the detailed reply!
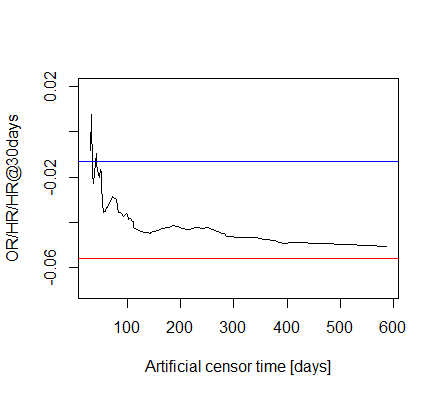
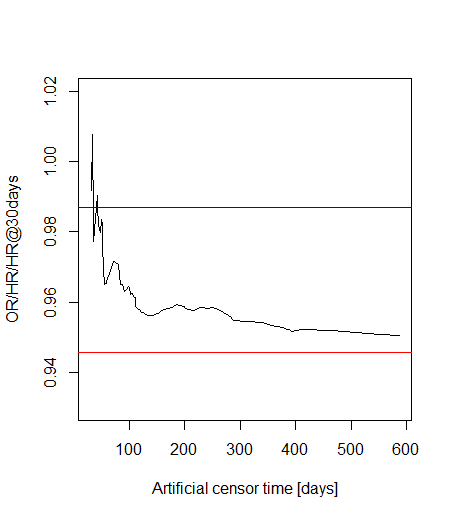
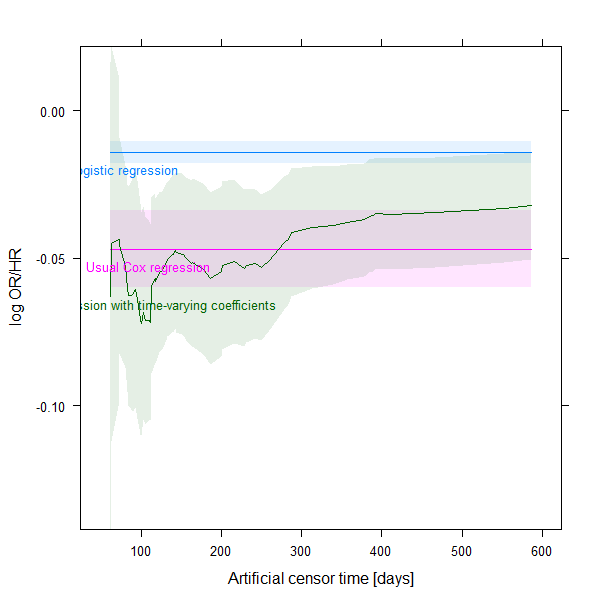
The fact that method 3 converges to something with increasing X is indeed nice, however, that method 2 and 3 converges to each other is not trivial (at least in my understanding!) as one represents a Cox regression with proportionality assumption, but it is relaxed in the other.
Ah, of course, sorry. I don’t want to introduce further transformations, but the vertical axis should indeed be relabeled.
There is another way to look this, and I really just forget to do it originally: we could also have a look at the confidence intervals! My updated code below will do this. (But I am not 100% sure in how to calculate the CI for the spline, hope this is correct…)
That’s right, perhaps the example is not the best from this aspect.
Well, we have 39 deaths for 3 covariates, that doesn’t seem horribly bad, but I now increased the cut-off time to 60 days, so we surely have plenty of events.
Updated code:
library( survival )
library( splines )
data( veteran )
veteran <- veteran[ veteran$time < 900, ] # outliers, likely coding error
veteran <- veteran[ veteran$status==1, ] # to simulate my (easier) situation when
# we have no censoring
vettimes <- sort( unique( veteran$time ) )
CutoffDay <- 60
# 1st approach: logistic regression
fit1 <- glm( I(veteran$time<=CutoffDay) ~ trt + prior + karno, data = veteran )
summary( fit1 )
coeffit1karno <- data.frame( times = vettimes[ vettimes>CutoffDay ],
approach = "Logistic regression",
coef = coef( fit1 )[ "karno" ],
t( confint( fit1 )[ "karno", ] ),
row.names = NULL )
# 2nd approach: censor everyone (artificially) at 30 days, and run a usual
# Cox-regression (i.e. proportionality assumption applies)
veteran$status2 <- ifelse( veteran$time<CutoffDay, 1, 0 )
veteran$time2 <- ifelse( veteran$time<CutoffDay, veteran$time, CutoffDay )
fit2 <- coxph( Surv( time2, status2 ) ~ trt + prior + karno, data = veteran )
summary( fit2 )
coeffit2karno <- data.frame( times = vettimes[ vettimes>CutoffDay ],
approach = "Usual Cox regression",
coef = coef( fit2 )[ "karno" ],
t( confint( fit2 )[ "karno", ] ),
row.names = NULL )
# 3rd approach: censor everyone (artificially) at X days (x>30), and run a
# Cox-regression with interaction with time (i.e. proportionality assumption
# relaxed).
fit3s <- lapply( vettimes[ vettimes>CutoffDay ], function( t ) {
veteran$status2 <- ifelse( veteran$time<t, 1, 0 )
veteran$time2 <- ifelse( veteran$time<t, veteran$time, t )
veteran3 <- survSplit( Surv( time2, status2 ) ~ trt + prior + karno,
data = veteran, cut = 1:max( veteran$time ) )
coxph( Surv( tstart, time2, status2 ) ~ trt + trt:ns( time2, df = 4 ) +
prior + prior:ns( time2, df = 4 ) +
karno + karno:ns( time2, df = 4 ), data = veteran3 )
})
coeffit3karno <- do.call( rbind, lapply( fit3s, function( f ) {
SplineMat <- cbind( 1, eval( attr( terms( f ), "predvars" )[[ 4 ]],
data.frame( time2 = CutoffDay ) ) )
sel <- grep( "karno", names( coef( f ) ) )
se <- sqrt( diag( SplineMat%*%vcov( f )[ sel, sel ]%*%t(SplineMat) ) )
data.frame( approach = "Cox regression with time-varying coefficients",
coef = SplineMat%*%coef( f )[ sel ],
X2.5.. = SplineMat%*%coef( f )[ sel ] + qnorm( 0.025 )*se,
X97.5.. = SplineMat%*%coef( f )[ sel ] + qnorm( 0.975 )*se )
} ) )
# Comparing the three approaches
coeffitskarno <- rbind( coeffit1karno, coeffit2karno,
data.frame( times = vettimes[ vettimes>CutoffDay ],
coeffit3karno ) )
Hmisc::xYplot(Hmisc::Cbind( coef, X2.5.., X97.5.. ) ~ times,
groups = approach, data = coeffitskarno, type = "l",
ylab = "log OR/HR", xlab = "Artificial censor time [days]",
method = "filled bands",
col.fill=scales::alpha(lattice::trellis.par.get()$superpose.line$col,
0.1 ) )
And the results: