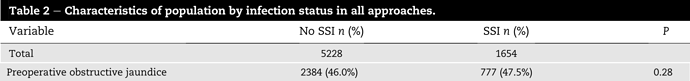
So I’m doing a meta-analysis for the first time and I’m trying to get through some new (to me) statistical techniques using R. To test whether I’m used the code correctly, I’ve been using a few of the studies absolute data and calculating the stats and checking if I get the same results as reported in the studies. However, I keep getting slightly different results and I’m wondering why.
Is this normal? Using the same test but different software and getting slightly different results?
For Example:
When I use R, I get a RR of 0.51 (0.36-0.72) using the code below:
library(epiR)
RiskFactor ← matrix(c(28, 103, 253, 351), ncol=2, byrow = TRUE)
colnames(RiskFactor) ← c(‘Infection’, ‘NoInfection’)
rownames(RiskFactor) ← c(‘LiverDis’, ‘NoLiverDisease’)
RiskFactor ← as.table(RiskFactor)RiskFactor
epi.2by2(RiskFactor, method=“cohort.count”)
Could I ask a few of you experts to use your stats tool of choice to compare?