Hi there,
I create this topic because this is a problem that I am facing now when validating my Cox PH model with validate() function.
My dataset consists in 666 observations, 501 events and 27 covariates (10 categorical and 16 continous).
Once I have developed a model and I want to validate() and calibrate() it, I find problems because there are three categories of a categorical covariate that have few observations (AH, MEMT and X).
e.g. X matrix deemed singular for covariate FLUID2 value X
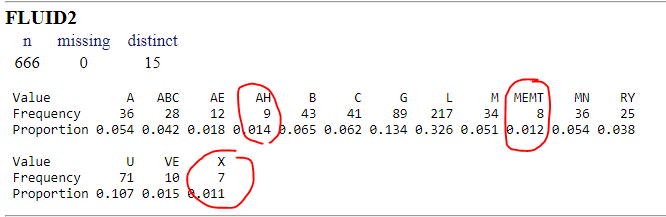
Although I did my best in reagrouping the categories I still have problems to run a boostrap without issues.
An idea that came to my mind is to perform the bootstrap avoiding the observations with those FLUID2 categories and keep the main model with them (considering these FLUID2: AH, MEMT and X observations). But I do not know if it would be correct or not.
As well, I do not know if there are better ways to manage these issue that for sure anyone of you have faced before. Any help will be highly appreciated.
Very best regards and many thanks for your support,
Marc