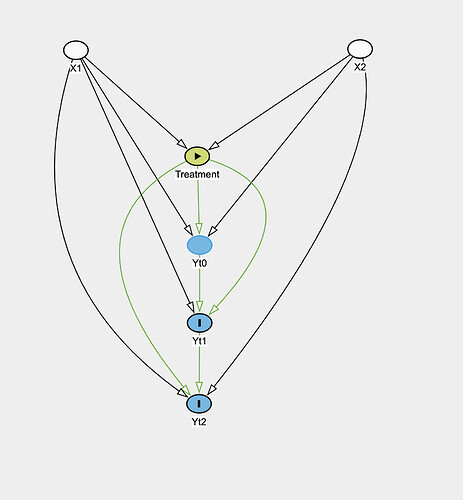
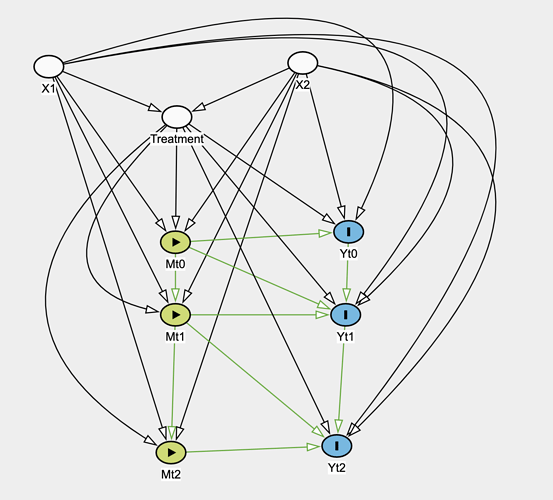
So take a situation in observational data where you have time-fixed treatment T, time-fixed covariates X_i, outcome Y_it. Treatment is not randomized and directly depends on covariates, which also influence the outcome. The following is the causal graph:
Essentially the covariates and treatment affects each of the outcomes which are also influenced by previous outcome. How would you estimate the causal effect of the treatment?
I’m new to graphs and in Biostat learned things like Mixed Models, ANCOVA, etc but the newer causal inference graphical model SEM stuff seems to be suggesting these approaches are not really valid (or I haven’t made the connection from DAG to these).
I would like to find the causal effect of the treatment on the outcome at each time point. The daggity software is telling me that when looking at the effect of Treatment on Yt1 that adjusting for Baseline is adjusting for a mediator and not good for estimating the total effect of Treatment on Yt1, but that this adjustment is needed to estimate the “direct” effect.
In school, we also learned that you could just do a mixed model and treat (Yt0,Yt1,Yt2) as the outcome, do a random intercept for Subject ID and then Treatment, X as covariates. Then I found out about Fixed Effects and how it is better to include Subject ID as a fixed effect so as to account for unobserved time fixed confounders: Fixed effects model - Wikipedia
However the fixed/mixed effects approach (with all Ys as the outcome) still wouldn’t consider the possible effect of the previous outcome on the current right? That is what this paper seems to suggest with assumption (b): https://imai.fas.harvard.edu/research/files/FEmatch.pdf.
Whereas doing a model for Yt1 against Yt0, X, Treatment would? And then another model for Yt2 vs Yt1, X, Treatment? That seems to be what the graph suggests would give the direct effect of the Treatment on the outcome at each time point.
I am just really confused with these newer causal graph techniques and how they relate to what I already learned, because prior to learning about them I probably would have just thrown this sort of data into a mixed model and looked at the Treatment coefficient. But now I am realizing that may not be correct…
Attached is the causal graph. What model would be reasonable for estimating the treatment effect with this graph?