Hi all!
I have one question about analysis of series of n-of-1 trials (i.e., a repeated crossover trial) with a continuous outcome (post-prandial glucose level in mg/dL). In general terms, I am wondering how to check if the variation of individual treatment effects can be at least partially “explained” by a baseline covariate. More details are below.
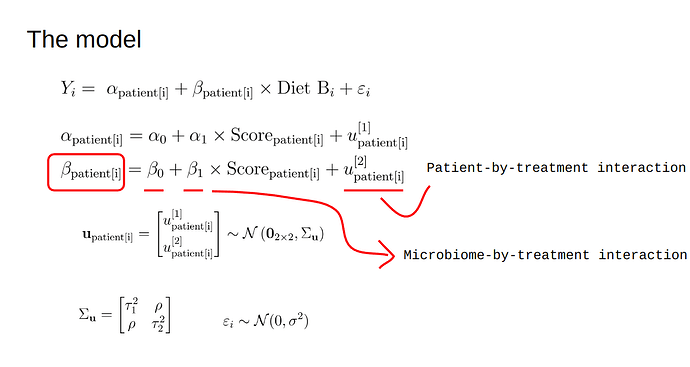
The plan is to use a mixed-effects model and assess the Heterogeneity of Treatment Effects at the individual level (diet A vs diet B for each patient, 5 crossover repeats per patient). However, we also want to assess the impact of a baseline covariate (a microbiome-related score) on this individual treatment effect. A simplified version of the model is shown below
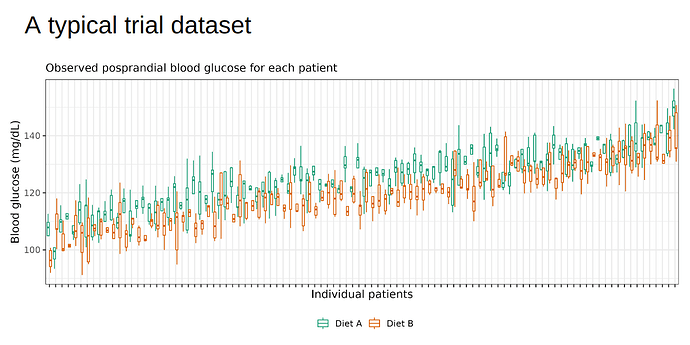
In words, each patient has its own intercept (alpha) and treatment effect (beta) (i.e., random intercepts and slopes), both of which also depend on the baseline microbiome score. A typical (simulated) dataset would look like below
Is this a reasonable strategy?
I know the baseline score for the intercept is redundant given the patients’ random intercepts (ie, \alpha_1 is redundant due to u^{[1]}_{\textrm{patient}[i]}), but I wonder if there’s any other way to estimate the dependency of the individual treatment effect on the microbiome score (\beta_1). I ran some simulations which do seem to work well, but any further guidance would be much appreciated.
Thank you very much!
Best,
Giuliano
2 Likes
My initial impression is that this study may be trying to make up with breadth (i.e., enrollment) what it lacks in depth. What if you enrolled just 10 highly motivated participant-collaborators, and studied their postprandial blood glucose more intensively — say, with CGM? What if you varied each participant’s diet adaptively, in response to evolving [individual-level] hypotheses about impact of diet on blood glucose?
As you are currently approaching the problem, it looks more like an exploration of hierarchical modeling per se than of any patient-centered scientific hypotheses about optimizing diet.
2 Likes
Thanks a lot for the feedback!
We are indeed considering continuous monitoring, but still without confirmation. The main research questions we are addressing are:
1. Is there variation in individual diet effects?
While this seems “obvious”, this has been claimed under inappropriate designs, including observational cohorts, parallel trials, and (non-repeated) crossover trials. None of these trials can identify patient-by-treatment interactions (as in Senn 2015). So, in that sense, our primary objective is intentionally “humble”. This has also to do with some microbiome hype/hope we are trying to “sort out”.
2. If existing, does the microbiome have any influence on this variation of IDEs?
Same justification as above.
Your idea about adaptively changing individuals’ diets is definitely the best approach as I see. To my knowledge, there are limited methods for that so far (beyond actually running the N-of-1 trials with multiple (>2) treatments, but that’s not adaptive). There may need to be some more methodological work along the lines of Shrestha & Sonia, 2021. Are you aware of any example trials doing something like that?
As for the number of patients, that was because we are not expecting huge variations in IDEs, so we wanted to make sure any variation is well estimated.
1 Like