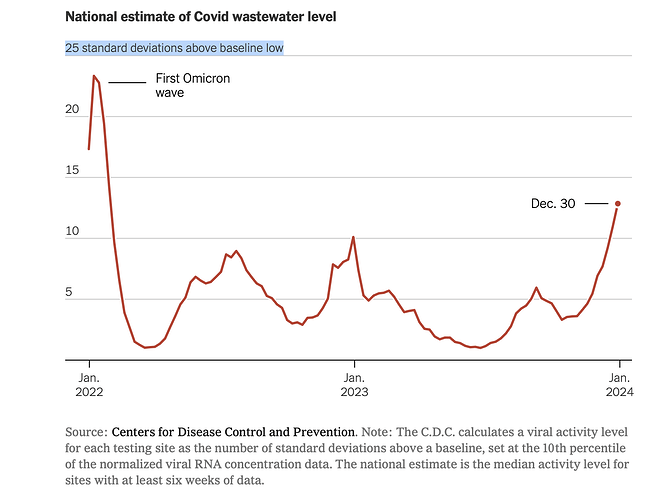
This chart from The New York Times free link caught my eye. The x-axis is time and the y-axis is the number of standard deviations above a baseline low, defined as the 10th percentile. The purpose is to estimate trends in COVID-19 prevalence based on wastewater sampling.
This appears to be a different approach than the cited paper.
I am a dilettante. My interest in applied statistics is exceeded only by my fear of appearing ridiculous. So, before sharing my incredulous reaction (“it’s cargo cult science”), your reactions would be helpful.
Following links from the CDC’s most helpful page I’m also bothered by
- Presence of viral RNA is established from three positive droplets measured by RT-qPCR or one when multiple assays or multiple PCR replicates are run. This seems skimpy.
- Normalization may be based on estimates of the served population. From past personal domain knowledge utilities did not collect information at the household level.
- A minimum of only 3 data points is required for trend estimation.
- Trends are identified by the slope of the linear regression, which ignores time series autocorrelation issues in regressing observations of log transformed SARS-CoV-2 normalized concentration against “date”
- Observations showing no virus are recorded as half the detection level.
This is even before aggregating results based on median values.
The methodology doesn’t pass my laugh test. Honestly, though, I’d rather be wrong than think that anyone is making public health decisions based on the results.
Am I being that guy who ______ ?
I’ve posted a longer take here .
The short short is that it is bitter to reflect on the public debate on the science during the pandemic when so much hostility was directed at the public health establishment for fake science that so many deaths ensued as a result of ignoring scientifically informed public health. Even though it appears that some of the initial advice was based on unexamined conventional wisdom discounting aerosol transmission, evidence and analysis is how science progresses.
There are also noble failures that chased down plausible lines of inquiry and learned little useful. Those, too, can be forgiven if anything at all can be salvaged from the wreckage.
Sewergate is the hardest pill to swallow. I read it as an example of Feynman’s Cargo Cult Science, at least as it is described on the CDC sources that I found. It would serve as a powerful, we told you so, you can’t trust government science bloody shirt. Taken on its face, the existence of a facially deficient analysis such as this has far more potential for harm arising from discrediting science-informed policy making than it could possibly be offset by the tea-reading it offers.