I am interested in the prognostic value of a biomarker for a complication following surgery. The complication is diagnosed using a continuous lab value (which in diagnostic criteria is dichotomized, but we can analyze as a continuous variable) measured during the 48 hour period following surgery. For the sake of this post, I’ll call the biomarker Z. This marker takes on a continuous value, typically between 0 and 1. We usually measure Z at two time points: immediately prior to surgery (when the complication has presumably not yet occured) and immediately after surgery (when the complication has presumably occurred, although the lab value used to diagnose the injury won’t reflect this until 1-2 days later).
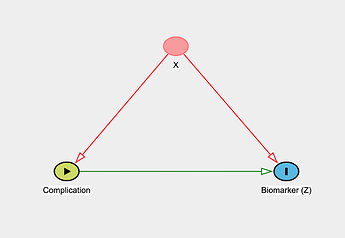
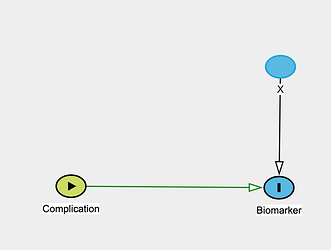
The goal here is to understand if Z has value in predicting the occurrence of this complication (which is officially ascertained 1-2 days after surgery using the standard lab value as noted above). Notably there are a few factors that we know can alter the level of the biomarker irrespective of the complication (call these X). These factors, though, are also documented risk factors for the complication itself (simple DAG shown below).
My team has discussed a number of ways to approach modeling the relationship between Z and the complication, and at this point I think I’ve run my mind in circles to the point where I’m not sure how best to proceed. I’ve outlined a bit more detail on the dataset below as well as unanswered questions. I would be very grateful for any input you all may have!
Dataset: ~800 observations (this is composed of patients with both pre- and post-surgery values for Z, and those with either only pre- or post-values; this is retrospective data).
Proposed outcome: peak 48-hour continuous lab value (adjusted for baseline pre-surgery value)
Biomarker details: when measured at either pre- or post- timepoints, there are variable repeated measures for each patient (ranging from 1-3 values).
Unanswered questions:
- It seems it would be reasonable to build this as a multi-level model (clustered by patient) given the repeated measures of Z at any given time point, is that correct?
- How should the pre- and post-surgical timepoints be incorporated? Does the imbalance in patients with values at pre-, post-, or both timepoints influence this choice?