What I’ve settled on after a lot of thought is the following:
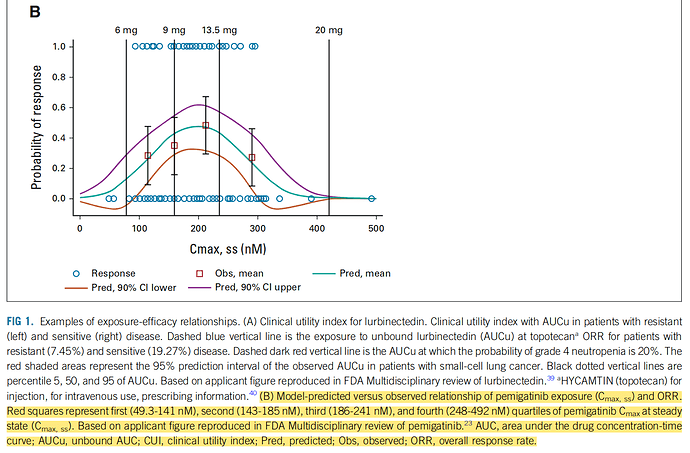
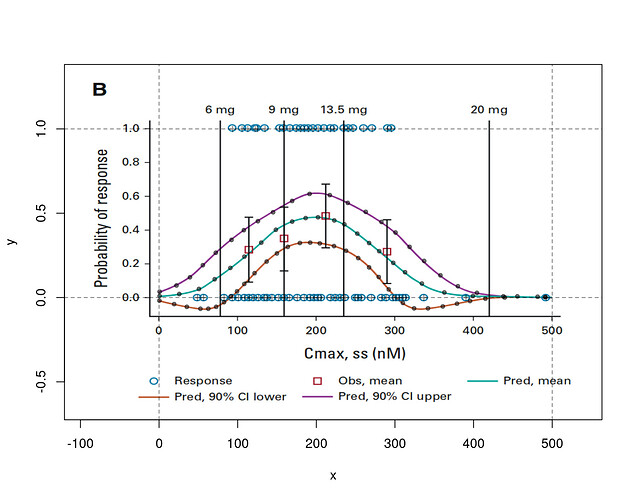
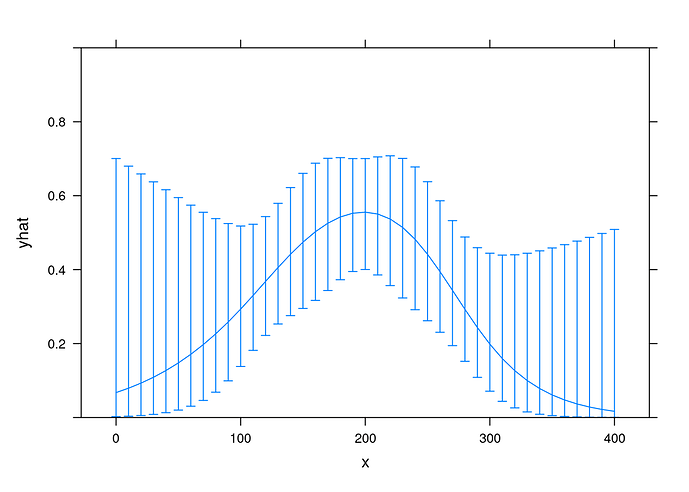
Here’s a plot exhibiting registration of my digitization of the data, using R package digitize
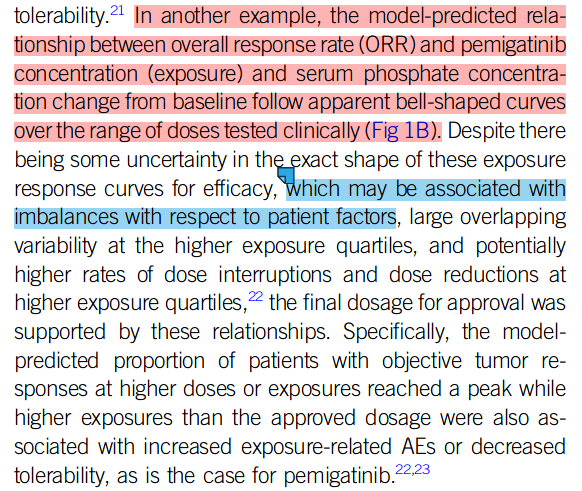
The original figure came from p 192 of this FDA CDER Multidisciplinary Review, which states, “The relationship followed a bell-shaped curve and a binary logistic regression with quadratic function was used to evaluate the relationship.”
library(rms, quietly = TRUE)
fit <- lrm(y ~ poly(x,2), data = exposure_response)
xYplot(Cbind(yhat, lower, upper) ~ x
, data = Predict(fit, x = 10*(0:40), fun = plogis, conf.int = 0.9)
, type = 'l'
, ylim = 0:1
)
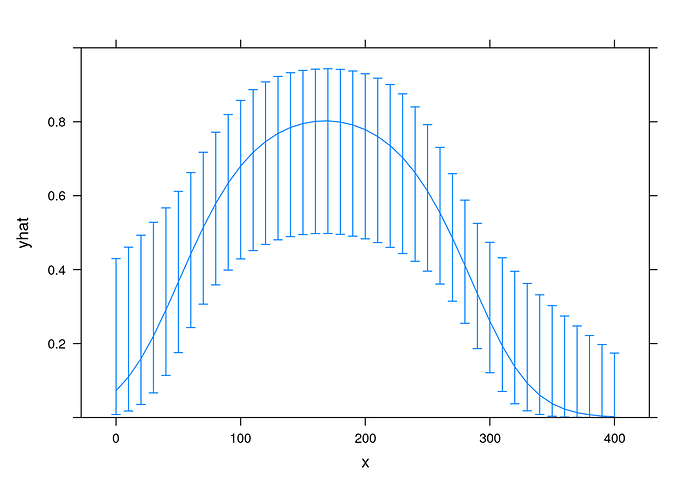
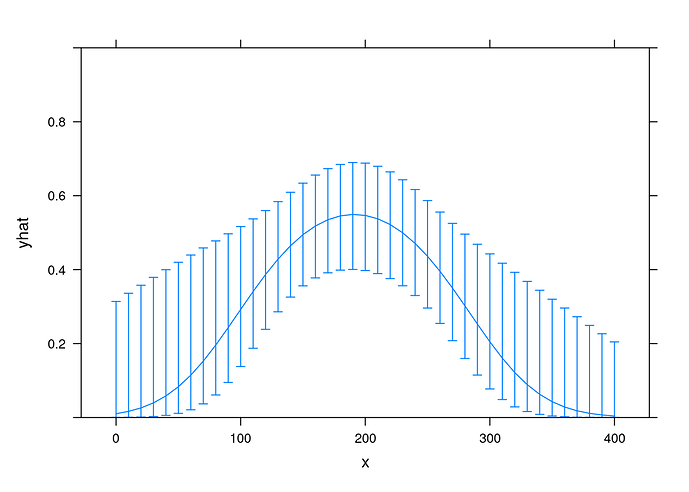
FYI, here is a spline regression for comparison:
fit2 <- lrm(y ~ rcs(x,4), data = exposure_response)
xYplot(Cbind(yhat, lower, upper) ~ x
, data = Predict(fit2, x = 10*(0:40), fun = plogis, conf.int = 0.9)
, type = 'l'
, ylim = 0:1
)
To evaluate the implied informational content of Figure 1B, I fitted Beta distributions to the estimated response probabilities using the method of moments — see e.g., BDA3 Eq (A.3), p583:
xout <- seq(0, 400, 25)
df <- data.frame(x = xout
, μ = with(meancurve, spline(x, y, xout=xout)$y)
, lcl = with(lclcurve, spline(x, y, xout=xout)$y)
, ucl = with(uclcurve, spline(x, y, xout=xout)$y)
)
## Add method-of-moments estimates; see BDA3 Eq (A.3), p. 583.
df <- Hmisc::upData(df
, stderr = (ucl - μ)/qnorm(0.95)
, `α+β` = μ*(1-μ)/stderr^2 - 1
, α = `α+β`*μ
, β = `α+β` - α
, print = FALSE
)
| x |
μ |
lcl |
ucl |
stderr |
α+β |
α |
β |
| 0 |
0.0088 |
-0.0179 |
0.0336 |
0.0151 |
37.4802 |
0.3308 |
37.1494 |
| 25 |
0.0204 |
-0.0451 |
0.0780 |
0.0350 |
15.2738 |
0.3115 |
14.9623 |
| 50 |
0.0493 |
-0.0654 |
0.1651 |
0.0704 |
8.4431 |
0.4160 |
8.0270 |
| 75 |
0.1224 |
-0.0500 |
0.2772 |
0.0941 |
11.1325 |
1.3630 |
9.7696 |
| 100 |
0.2099 |
0.0403 |
0.3714 |
0.0982 |
16.2103 |
3.4027 |
12.8076 |
| 125 |
0.3052 |
0.1613 |
0.4507 |
0.0885 |
26.0998 |
7.9654 |
18.1344 |
| 150 |
0.4066 |
0.2618 |
0.5199 |
0.0689 |
49.8290 |
20.2603 |
29.5687 |
| 175 |
0.4588 |
0.3196 |
0.5865 |
0.0776 |
40.2055 |
18.4459 |
21.7596 |
| 200 |
0.4761 |
0.3233 |
0.6190 |
0.0869 |
32.0648 |
15.2671 |
16.7978 |
| 225 |
0.4543 |
0.3016 |
0.5939 |
0.0848 |
33.4422 |
15.1935 |
18.2487 |
| 250 |
0.3868 |
0.2449 |
0.5391 |
0.0926 |
26.6580 |
10.3107 |
16.3473 |
| 275 |
0.2939 |
0.1437 |
0.4727 |
0.1087 |
16.5680 |
4.8694 |
11.6986 |
| 300 |
0.1974 |
0.0079 |
0.3887 |
0.1163 |
10.7113 |
2.1140 |
8.5973 |
| 325 |
0.1097 |
-0.0649 |
0.2703 |
0.0976 |
9.2513 |
1.0152 |
8.2361 |
| 350 |
0.0488 |
-0.0598 |
0.1622 |
0.0690 |
8.7612 |
0.4276 |
8.3336 |
| 375 |
0.0189 |
-0.0405 |
0.0789 |
0.0365 |
12.9046 |
0.2435 |
12.6611 |
| 400 |
0.0091 |
-0.0168 |
0.0327 |
0.0143 |
42.8959 |
0.3905 |
42.5054 |
To the extent that the \mathrm{Beta}(\alpha, \beta) can be interpreted as equivalent to \alpha-1 successes and \beta-1 failures in \alpha+\beta-2 Bernoulli trials (cf. BDA3 p.35), these calculations suggest that the plotted curves embody strong prior assumptions that go far beyond the data collected, and indeed beyond any data that ever could be collected, since you can’t observe fewer than 0 successes. Thus, the form of the analysis seems to have ‘put a thumb on the scale’. How reasonable does this mode of interpretation seem to the Bayesians here?